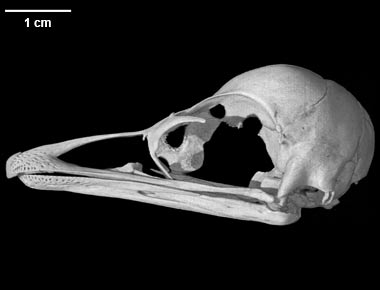
Dromaius novaehollandiae, the emu, is a paleognath bird. This is a group of large, flightless birds that also includes the extinct elephant bird and moa and the extant ostrich, rhea, kiwi, and tinamou. Emus are indigenous to Australia, and have recently been raised in captivity for their eggs, meat, and skin. Emus usually weigh between 50-55 kg, and have an average height of 1.75m, making them the second largest living bird. Like ostriches, emus are very fast, capable of top speeds around 50 km/hr; they are also strong swimmers. The emus diet generally consists of seeds, fruits, and young shoots, but they will eat insects and small vertebrates when available. (Information obtained from the University of Michigan Museum of Zoology page - see links).

About the Species
The specimen used for this project (TMM M-6757) is a juvenile. It was ranch-raised and donated by the Rockin' W Ranch in Dripping Springs, Texas.

About this Specimen
This specimen was scanned by Matthew Colbert on 05 February 2001 at the University of Texas High-Resolution X-ray CT Facility. The animations and slices for this project were made using the following software programs: Adobe Photoshop 6.0, NIH Image 1.62 (with CT Macro_Mac file), File Buddy 6.05, Illustrator 9.02, and QuickTime Movieplayer Pro.
The first step in preparing the images was to make them visible, and to change them from 16-bit to 8-bit format. The Input values used for the levels were 0, 1, 15, and the mode was subsequently changed from 16-bit to 8-bit. The images were then resliced vertically and horizontally in NIH Image - producing sagittal and horizontal slice planes respectively. The interslice distance used for the Input spacing was 2.31 pixels. This was calculated by: (Image dimension of 512 pixels/Field of reconstruction of 30.3mm) X Interslice spacing of 0.137mm. The Output spacing used was 1 pixel. The coronal images were cropped to reduce file size and processing time before reslicing. After reslicing, the horizontal plane consisted of 414 slices, and the sagittal plane consisted of 408 slices; the original coronal plane also had 408 slices. These numbers included 1 blank slice at the front and back of each slice plane; additional blank slices were deleted. The sagittal and horizontal stacks were also rotated to orient them in a similar fashion to the coronal slices (dorsal surface up, ventral surface down, snout facing the same direction when possible, etc.). The interslice spacing of the new slice planes was 0.059mm. A final leveling pass was then done in Photoshop to reduce noise, and to make the bone easier to see. The Input grayscale values used were 70, 1, 200, and the Output numbers were 0, 239.
Next, the images had to be numbered. In order to number the files in NIH Image, the file names had to be changed, and this was done in File Buddy. The Basename was left empty, the Start Numbering was set at 001, the Increment was kept at 1, and the Append to each name was left empty. An extra step was done when renaming the sagittal plane; the stack was flipped with the triangle button such that the last slice became slice number one, and the first slice became the last slice, because NIH Image causes the sagittal slice plane to be inverted. Once renamed, the images were all reopened in NIH Image and numbered. The numbered files were then stacked, and saved as TIFF images. This was done for all three slice planes.
QuickTime movies were then made for all three slice planes. This was done by opening the TIFF images in Movieplayer Pro. In Open Image Sequence, a frame rate of 15 frames per second was chosen. The movies were then Exported and saved with a .mov extension. The exported settings were: Compression: Graphics; Depth: Grayscale; and Quality: Best. 15 was put as the frame rate again.
The last thing that needed to be done was to make files that could be labeled. This was done by going back to the 8-bit, unnumbered, but completely leveled images. Canvas sizes were experimented with to see how much space had to be added for labeling. The canvas was then added, resulting in an image size of 7" X 7", or 504 pixels X 504 pixels. After batching the canvas addition, the images were numbered in NIH as described above. The images were now ready to be labeled in Illustrator. The labeling conventions used were as follows: Document Color Mode and all labels- RGB color. Font size- 14 pt, Font type- Arial Regular, Line width- 2 pixels with a shadow. Bone- Pure red (255, 0, 0) with all capitalized abbreviations. Processes, passageways, open spaces, and foramina- Pure yellow (255, 255, 0) with all lower case abbreviations. Foreign substances (such as wax)- Pure blue (0, 0, 255) with all lower case abbreviations. Each color was put in a different layer, and the labeled images were then saved as Illustrator files. These labeled images will be made available here once I complete my research.

About the
Scan
Literature
Baumel, J. J. (ed.). 1993. Handbook of Avian Anatomy: Nomina anatomica avium, Second edition. Publications of the Nuttall Ornithological Club. 779 pp.
Heilmann, G. 1972. The Origin of Birds. Dover Publications, Inc. New York. 209 pp.
McDowell, S. 1948. The bony palate of birds. Part I. The Palaeognathae. The Auk 65:520-549.
Parker, W. K. 1866. On the structure and development of the skull in the ostrich tribe. Philosophical Transactions of the Royal Society of London 156:113-183.
Proctor, N. S., and P. J. Lynch. 1993. Manual of Ornithology, Avian Structure and Function. Yale University Press, New Haven, CT. 340 pp.
Webb, M. 1957. The ontogeny of the cranial bones, cranial peripheral and cranial parasympathetic nerves, together with a study of the visceral muscles of Struthio. Acta Zoologica 38:81-203.
Zusi, R. 1993. Patterns in diversity in the avian skull; pp. 391-437 in J. Hanken, and B. K. Hall (eds.); The Skull, Volume 2- Patterns of Structural and Systematic Diversity. The University of Chicago Press. 566 pp.
Links
Dromaius novaehollandiae on The Animal Diversity Web (The University of Michigan Museum of Zoology)

Literature
& Links
None available.

Additional
Imagery
|